MAPP (Mass Spectrometry Analysis of Proteolytic Peptides) is a groundbreaking technology that isolates and identifies proteins actively degraded by the proteasome, offering an unparalleled and direct view of the "active degradome." This innovative approach provides a sensitive and comprehensive analysis of degradation products, distinguishing it as the only technology capable of examining proteasome-mediated degradation directly. MAPP holds transformative potential in identifying novel therapeutic targets, such as differentially degraded or stabilized proteins under varying conditions or disease states. Its applications span various human diseases, including cancer, autoimmune disorders, infections, and beyond, paving the way for breakthroughs in precision medicine and targeted therapies.
- Target Discovery: Reveal novel proteins differentially degraded or stabilized under specific disease conditions and identify disease-specific degradation networks/pathways relevant to disease mechanisms, offering insights into new therapeutic targets and enhancing tumor antigenicity or immune recognition.
- Biomarker Discovery and precision medicine: Identify degradation-derived peptides as potential biomarkers for disease progression or therapeutic response. Identifies proteasomal degradation "fingerprints" in biopsies, tumor samples, and biological fluids, aiding personalized treatment selection across different diseases, including cancer, autoimmunity, neurodegeneration, and infectious diseases.
- Efficacy Studies in PROTAC-Related Research: stratifying patients, examining downstream effects, and comparing different lead compounds.
- High Sensitivity: Detects low-abundance proteins and proteins with rapid turnover that traditional methods may miss.
- Unbiased Sampling: Provides an unbiased view of the active degradome in single-peptide resolution.
- Versatile: Applicable to various sample types and biological contexts, from basic research to clinical studies.
- Novel Insights: Offers a unique perspective on protein cleavage and regulatory steps, enhancing understanding of aberrations in proteostasis, degradation events, and immune presentation
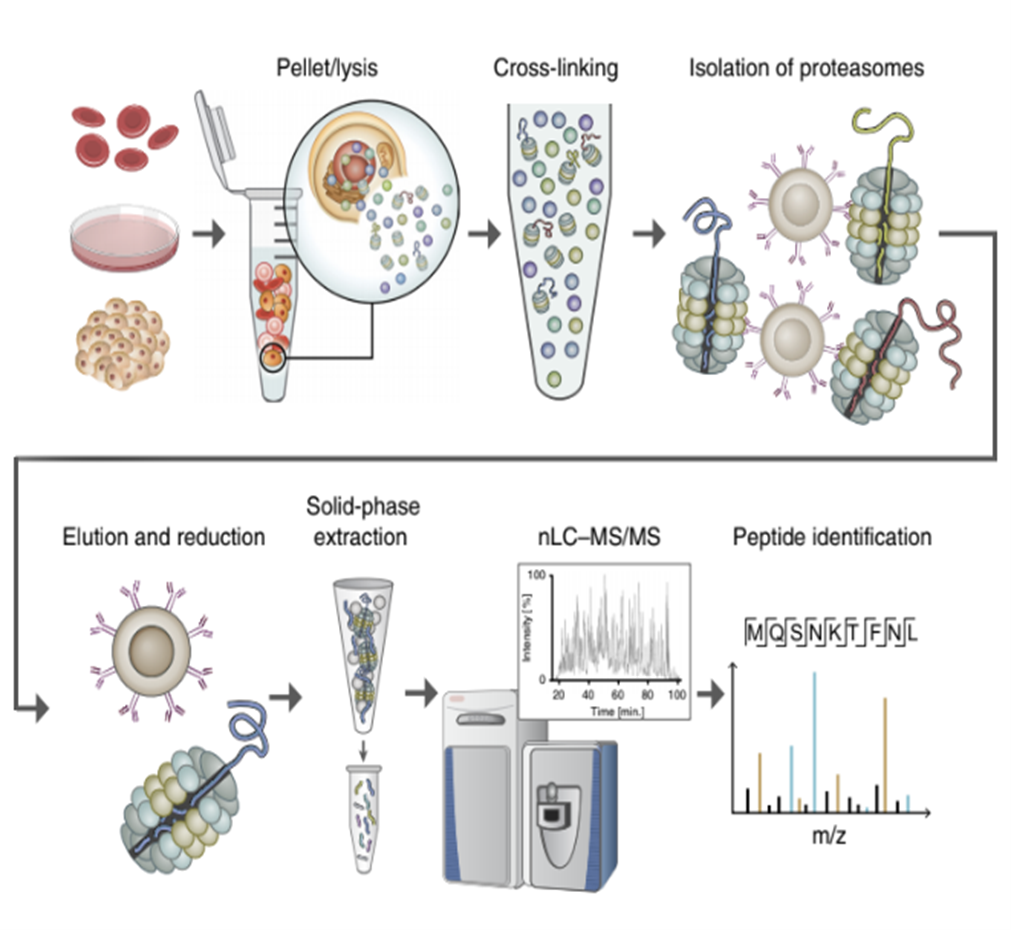
MAPP method workflow
Javitt, A., Shmueli, M.D., Kramer, M.P. et al. The proteasome regulator PSME4 modulates proteasome activity and antigen diversity to abrogate antitumor immunity in NSCLC. Nat Cancer 4, 629–647 (2023).
Wolf-Levy H, Javitt A, Eisenberg-Lerner A, Kacen A, Ulman A, Sheban D, Dassa B, Fishbain-Yoskovitz V, Carmona-Rivera C, Kramer MP, Nudel N, Regev I, Zahavi L, Elinger D, Kaplan MJ, Morgenstern D, Levin Y. Protocols demonstrating MAPP's sensitivity in distinguishing systemic lupus erythematosus patients from healthy donors. Nat Biotechnol. 2016; 540:544–551.


