Prof. Ron Milo and his team have engineered a bacteria (E. coli) to fix atmospheric CO₂, converting it into organic biomass. This breakthrough synthetic biology approach enables E. coli to use CO₂ as its sole carbon source by harnessing energy from formate, offering a potential solution to the major global challenge of reducing atmospheric CO₂ levels and a potentially unique platform for precision fermentation.
- Potential Precision Fermentation Platform: A modular platform for producing various chemicals with net negative CO₂ emissions by integrating synthetic metabolic pathways.
- Research Tool: Useful for studying and improving enzymes in the Calvin-Benson-Bassham (CBB) cycle, contributing to advancements in crop efficiency.
- Sustainable: Provides a way to sequester atmospheric CO₂
- Genetic Engineering Compatibility: E. coli-based standard laboratory techniques can be applied for genetic modification.
The Milo team has developed and characterized carbon-fixing strains of E. coli, demonstrating autotrophic growth through continuous laboratory evolution. These strains have been validated via carbon economy assessments and genomic sequencing to identify the mutations that support autotrophy.
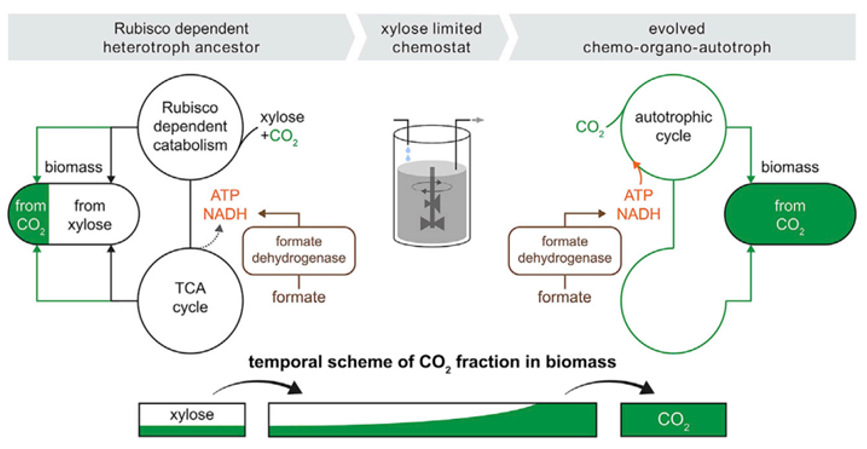
Figure 1: The experimental workflow begins from the left, where the E. coli strain is engineered for xylose catabolism (pfkA, pfkB, Zwf deleted; Rubisco, Prk, CA, FDH overexpressed) but is unable to grow in autotrophic conditions. As the limited amount of xylose is consumed in the chemostat, and with an excess of formate and CO2, the cells are under a strong selection pressure to use CO2 as the only carbon source while using formate oxidation by FDH as the energy source. Evolved clones with a fully autotrophic phenotype were then produced (right), as they achieved a fitness advantage over xylose-dependent strains and became the dominant population in the chemostat.


